SynErgie | Prognose-Service¶
Der Prognose-Service wurde in SynErgie vom Institut für Produktionsmanagement, Technologie und Werkzeugmaschinen (PTW) der TU Darmstadt entwickelt, um Ingenieur*innen die Erstellung von Prognose-Modellen der Nutzenergiebedarfe (Wärme, Kälte, Strom) in Produktionsbereichen zu erleichtern. Mit dem Service können die erstellten Modelle zusätzlich in Echtzeitumgebungen eingesetzt werden. Damit wird die prädiktive Regelung der Energieversorgungssysteme für den energieflexiblen Fabrikbetrieb unterstützt. Der Service soll Forschungspartner*innen und Unternehmen dabei unterstützen, den energieflexiblen Betrieb umzusetzen.
Der Service besteht, wie in Abbildung 1 gezeigt, aus den Schritten Modellierung (Teil 1) und Deployment (Teil 2). Die Nutzenergiebedarfsprognosen werden als Eingangsgröße für die modell-prädiktive Regelung der Produktionsinfrastruktur benötigt und können darüber hinaus für weitere Anwendungsfälle genutzt werden.
Die Einbettung des Prognose-Service in den Regelkreis der energieflexiblen Fabrik wird in Abbildung 1 aufgezeigt. Die Prognose bildet darin einen integralen Bestandteil, um die Energieversorgungssysteme vorausschauend betreiben zu können. Typische Eingangsgrößen für die Prognosemodelle sind Messdaten (Energiebedarf der letzten Zeit), Produktionsplandaten (geplante Fertigungsaufträge, Fabrikauslastung) und Wetterdaten (Wetterprognose des nächsten Tags). Mithilfe von aufgenommenen Daten wird im Schritt Modellierung ein Machine-Learning-Modell antrainiert. Dieses liefert dann nach dem Deployment im Live-Betrieb Prognosen für den Lastgang des Produktionssystems bis zu 24h in die Zukunft.
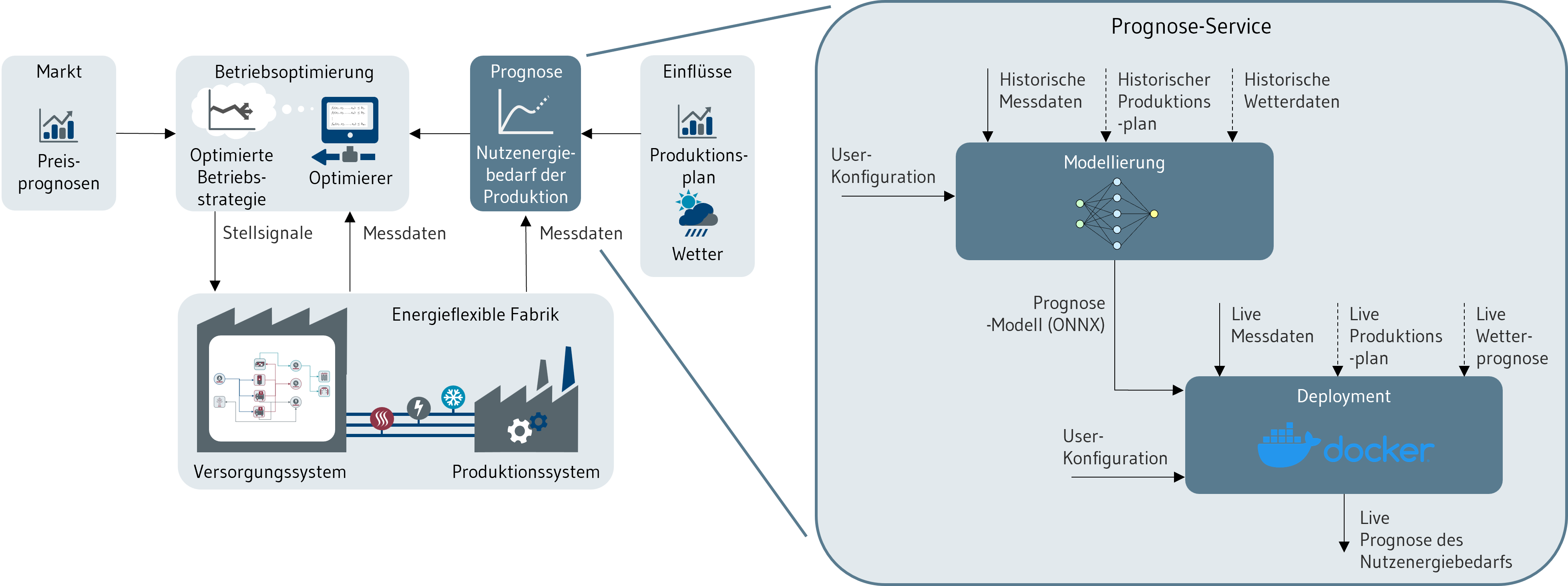
Abbildung 1: Einbettung des Prognose-Service in den Regelkreis der energieflexiblen Fabrik
Das Jupyter Notebook unten ist eine kurze Demonstration des Modellierungsteils des Services. Aus Sicherheitsgründen wird die Demonstration statisch zur Verfügung gestellt. Falls Sie SynErgie-Partner sind und den Service mit eigenen Daten testen möchten, wenden Sie sich bitte an die verantwortliche Person. Diese, sowie die Dokumentation des Services, findet sich unter folgendem Link: Dokumentation
Ein Video der Demonstration des Teils Deployment ist auf Youtube zu finden.
Video Prognose-Service
Im folgenden Video wird die Nutzung des Jupyter Notebooks erläutert. Unten können Sie in den Code-Zellen folgen.